Traditional microtomy relies on a fully manual cut-float-mount workflow that can take up to 2 hours per slide. The sections produced vary in thickness by ± 5 µm, retain only about 15 % of nuclei in 20-µm slices, and provide no accurate 3-D spatial reference for locating tissue structures.
Current diagnostic approaches still face major limitations: a 10–15% misdiagnosis rate persists across mainstream methods. Imaging tools such as X-ray, CT, and MRI provide limited molecular-level insight, while blood assays and PCR often lack sufficient biological context. Meanwhile, tissue pathology remains low in automation and throughput, frequently capturing only fragmented pieces of information.


Critical information is often lost in current workflows because 2D tissue slices discard a large amount of biological detail. Even adjacent sections can differ substantially, showing up to 50% variability in cellular morphology, which makes interpretation inconsistent. Without reliable 3D reconstruction, truly holistic analysis remains out of reach.
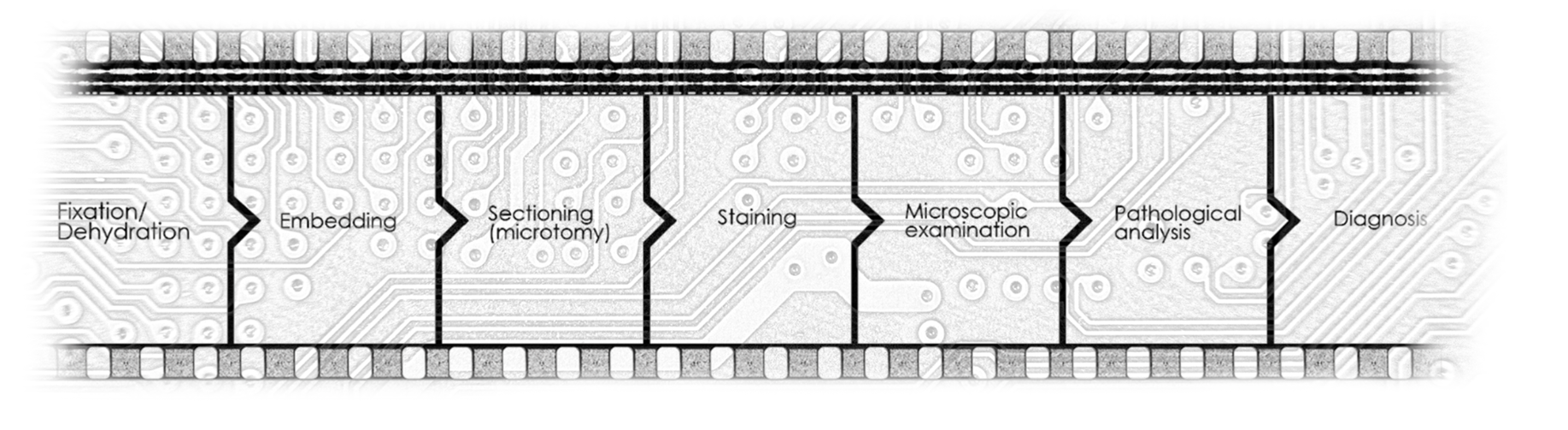
A tissue sample’s path through the lab has long been defined by fragmentation. Fixation, embedding, sectioning, staining, microscopic examination, and pathological analysis are typically handled as separate steps, with repeated handoffs that add time, complexity, and information loss.
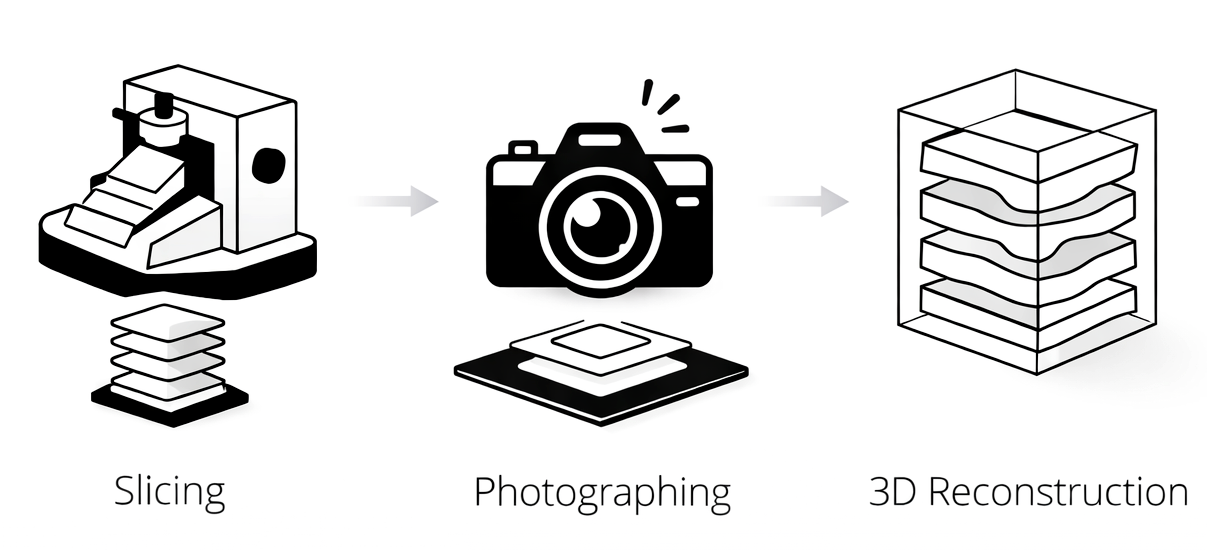
We integrate this entire chain into a single, fully automated, standalone system, creating a continuous workflow from preparation to interpretation. The shift starts at sectioning. With our proprietary AI at the core, the system automatically scans thousands of tissue slices and reconstructs them in 3D, transforming dispersed 2D fragments into a coherent volumetric view. This enables clearer visualization, streamlined review, and more precise localization of lesions, revealing where disease truly resides.

